🧭 Objective¶
🧠 What is Clustering?¶
📖 Click to Expand
🧠 What is Clustering?¶
Clustering is an unsupervised learning technique that groups similar data points together without predefined labels.
- The goal is to identify natural groupings in data based on similarity or distance
- Each group is called a cluster, and the points within a cluster are more similar to each other than to points in other clusters
- Used to uncover structure in data when no labels are available
Types of Clustering¶
- Hard Clustering: Each point belongs to one cluster (e.g., KMeans)
- Soft Clustering: Points can belong to multiple clusters with probabilities (e.g., GMM)
- Hierarchical Clustering: Builds a tree-like structure of nested clusters
- Density-Based Clustering: Forms clusters based on dense regions (e.g., DBSCAN)
📌 When is Clustering Useful?¶
📖 Click to Expand
📌 When is Clustering Useful?¶
Clustering is helpful when you want to discover structure or patterns in unlabeled data.
Common Applications¶
- Customer Segmentation: Group users based on behavior or demographics
- Market Research: Identify distinct buyer personas
- Anomaly Detection: Spot outliers as points that don’t belong to any cluster
- Recommender Systems: Group similar items or users
- Document Clustering: Group similar news articles, reports, etc.
- Genetics & Bioinformatics: Group similar gene expressions or cell types
Key Benefit¶
Clustering helps reduce complexity by summarizing large datasets into meaningful groups, even when labels are unavailable.
📏 Evaluation Challenges¶
📖 Click to Expand
📏 Evaluation Challenges¶
Clustering is difficult to evaluate because it’s unsupervised — there’s no ground truth.
Internal Evaluation¶
- Silhouette Score: Measures how well a point fits within its cluster vs. others
- Davies-Bouldin Index: Lower values = better cluster separation
- Calinski-Harabasz Index: Ratio of between- to within-cluster dispersion
External Evaluation (when ground truth is available)¶
- Adjusted Rand Index (ARI): Measures similarity to true labels
- Normalized Mutual Information (NMI): Captures mutual information between assignments and labels
Other Challenges¶
- Choosing the Number of Clusters (K)
- Handling High-Dimensionality: PCA/t-SNE often needed
- Scale Sensitivity: Many algorithms need feature normalization
- Irregular Cluster Shapes: Some methods fail on non-spherical clusters
📦 Data Setup¶
📥 Load Dataset¶
from sklearn.datasets import make_blobs
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
# Generate synthetic clusterable data
X, y = make_blobs(n_samples=500, centers=4, cluster_std=1.2, random_state=42)
df = pd.DataFrame(X, columns=["feature_1", "feature_2"])
# Plot raw input
plt.figure(figsize=(6, 4))
sns.scatterplot(data=df, x="feature_1", y="feature_2")
plt.title("Unlabeled Input Data")
plt.show()
🧹 Preprocessing¶
📖 Click to Expand
🧹 Preprocessing¶
Clustering algorithms like KMeans, DBSCAN, and GMM are sensitive to feature scale.
- We apply StandardScaler to normalize all features to zero mean and unit variance
- This ensures that distance-based calculations treat each feature equally
- Additional steps like missing value imputation or encoding are skipped here as all features are numeric and clean
from sklearn.preprocessing import StandardScaler
# Scale data
scaler = StandardScaler()
df_scaled = pd.DataFrame(scaler.fit_transform(df), columns=df.columns)
# Preview
df_scaled.head()
| feature_1 | feature_2 | |
|---|---|---|
| 0 | -0.766161 | 0.562117 |
| 1 | -1.266310 | -1.545187 |
| 2 | 2.006315 | -0.297524 |
| 3 | 0.077145 | 0.684294 |
| 4 | -0.385682 | -1.611826 |
📊 Clustering Algorithms¶
📊 Comparison¶
| Algorithm | Works Well For | Assumes Shape | Needs Scaling | Handles Outliers | Notes |
|---|---|---|---|---|---|
| KMeans | Spherical, equal-size blobs | Spherical | ✅ Yes | ❌ No | Fast, interpretable |
| Hierarchical | Any size, low dims | Flexible (linkage) | 🟡 Sometimes | ❌ No | Good for visualizing nested groups |
| DBSCAN | Irregular, density-based | Arbitrary | ✅ Yes | ✅ Yes | Needs eps tuning |
| GMM | Elliptical, probabilistic | Gaussian blobs | ✅ Yes | ❌ No | Soft assignments |
| Mean Shift | Smooth cluster shapes | Arbitrary | ✅ Yes | ❌ No | Bandwidth sensitive |
| Spectral | Graph-connected data | Graph-based | ✅ Yes | ❌ No | Slow for large N |
| OPTICS | Nested, variable density | Arbitrary | 🟡 Sometimes | ✅ Yes | Better than DBSCAN for chaining |
| BIRCH | Large data, streaming | Spherical-ish | 🟡 Sometimes | ❌ No | Memory-efficient |
| HDBSCAN | Hierarchical + density | Arbitrary | 🟡 Sometimes | ✅ Yes | Adaptive to density |
| Affinity Prop. | Message-passing structure | N/A (similarity) | ✅ Yes | ❌ No | No need to set K |
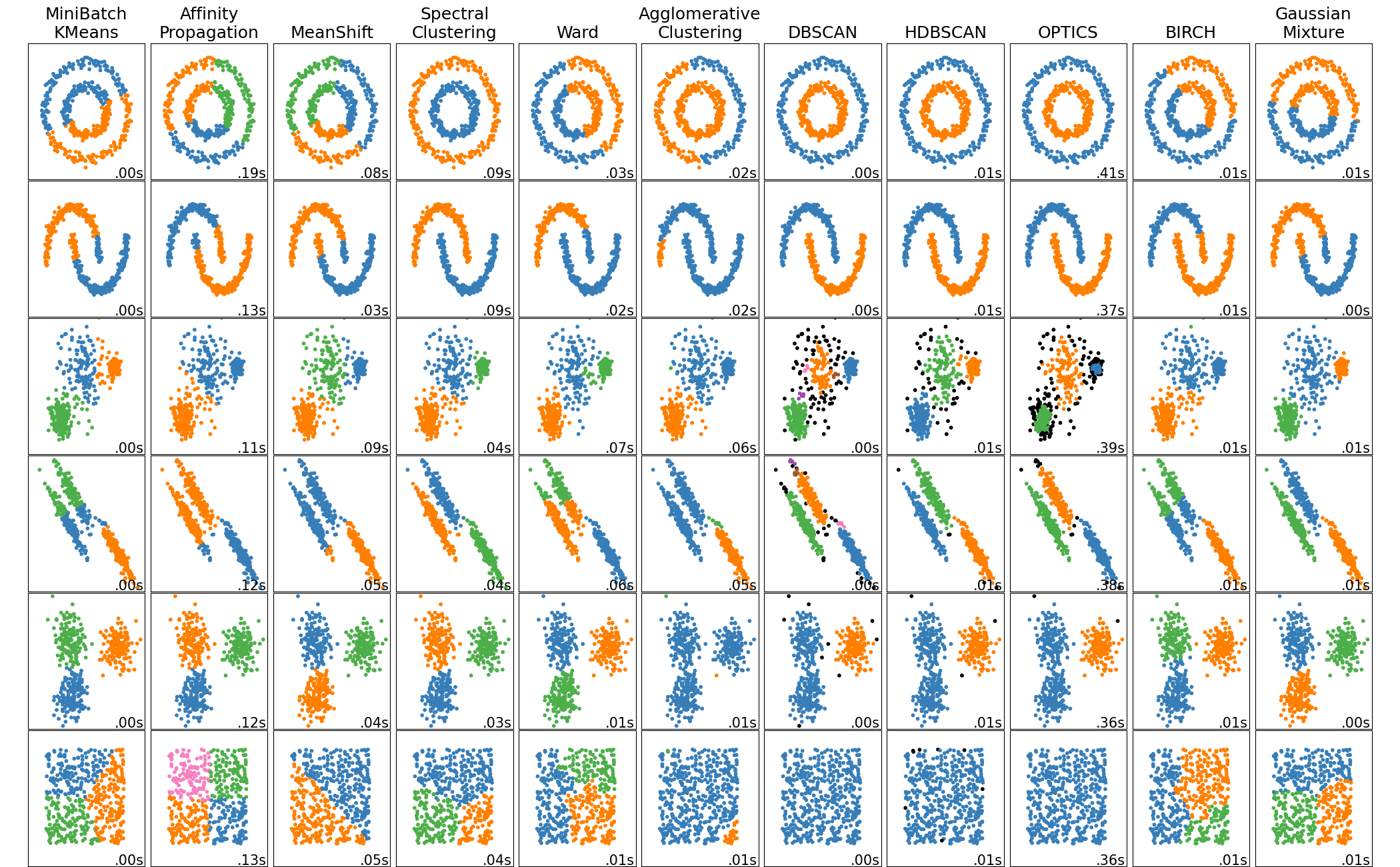
🖼️ Visual Comparison¶
This image from scikit-learn shows how different algorithms behave on varied shapes and densities:
📈 KMeans¶
📖 Click to Expand
📈 What is KMeans?¶
KMeans is a partitioning-based clustering algorithm that groups data into K clusters by minimizing the sum of squared distances (inertia) within clusters.
⚙️ How It Works¶
- Initialize
Kcentroids (randomly or via KMeans++) - Assign each point to the nearest centroid
- Update centroids as the mean of all assigned points
- Repeat steps 2–3 until convergence
✅ When to Use¶
- Clusters are compact, well-separated, and roughly spherical
- Scalability matters — KMeans is fast and efficient
- Applications like customer segmentation, image compression, etc.
⚠️ Limitations¶
- Requires choosing
Kin advance - Sensitive to initial centroids, outliers, and feature scaling
- Assumes clusters are convex and isotropic
🧠 Variants¶
- KMeans++: Better centroid initialization
- MiniBatch KMeans: Faster for large datasets
- Fuzzy C-Means: Soft assignment to multiple clusters
⚙️ KMeans Config¶
# 📈 KMeans — ⚙️ Config
from sklearn.preprocessing import StandardScaler
# === CONFIG ===
k_range = range(2, 10) # Range of K values to evaluate
k_final = 4 # Final K to use when fitting the model
random_state = 42 # Seed for reproducibility
use_scaled_data = True # Whether to scale before clustering
scaler = StandardScaler() # Scaler object
# Prepare input data
df_input = df.copy()
if use_scaled_data:
df_input = pd.DataFrame(scaler.fit_transform(df), columns=df.columns)
# Preview
df_input.head()
| feature_1 | feature_2 | |
|---|---|---|
| 0 | -0.766161 | 0.562117 |
| 1 | -1.266310 | -1.545187 |
| 2 | 2.006315 | -0.297524 |
| 3 | 0.077145 | 0.684294 |
| 4 | -0.385682 | -1.611826 |
📉 K Selection: Elbow + Silhouette¶
📖 Click to Expand
📉 K Selection Using Elbow + Silhouette¶
Choosing the right number of clusters (K) is critical to effective clustering.
🔹 Elbow Method¶
- Plots inertia (within-cluster sum of squares) vs. K
- Look for a point where the drop in inertia slows significantly — the "elbow"
- Simple and fast, but subjective
🔹 Silhouette Score¶
- Measures how well each point fits within its cluster vs. the next best
- Values range from -1 to 1:
- Closer to 1 → well-clustered
- Around 0 → overlapping clusters
- Negative → likely misclassified
- Useful for identifying over-clustering or under-clustering
🔁 Best Practice¶
Use both methods together:
- Elbow narrows the K range
- Silhouette refines the choice based on structure quality
from sklearn.cluster import KMeans
from sklearn.metrics import silhouette_score
import matplotlib.pyplot as plt
inertias = []
silhouettes = []
for k in k_range:
kmeans = KMeans(n_clusters=k, n_init=10, random_state=random_state)
labels = kmeans.fit_predict(df_input)
inertias.append(kmeans.inertia_)
silhouettes.append(silhouette_score(df_input, labels))
# Plot Elbow and Silhouette Score
fig, ax = plt.subplots(1, 2, figsize=(12, 4))
# Elbow plot
ax[0].plot(list(k_range), inertias, marker='o')
ax[0].set_title("Elbow Method (Inertia vs K)")
ax[0].set_xlabel("Number of Clusters (K)")
ax[0].set_ylabel("Inertia")
# Silhouette plot
ax[1].plot(list(k_range), silhouettes, marker='o', color='green')
ax[1].set_title("Silhouette Score vs K")
ax[1].set_xlabel("Number of Clusters (K)")
ax[1].set_ylabel("Silhouette Score")
plt.tight_layout()
plt.show()
🚀 Run KMeans¶
# Fit KMeans using the selected final K
kmeans_final = KMeans(n_clusters=k_final, n_init=10, random_state=random_state)
labels_final = kmeans_final.fit_predict(df_input)
# Append cluster assignments to the data
df_kmeans = df_input.copy()
df_kmeans["cluster"] = labels_final
df_kmeans.head()
| feature_1 | feature_2 | cluster | |
|---|---|---|---|
| 0 | -0.766161 | 0.562117 | 1 |
| 1 | -1.266310 | -1.545187 | 2 |
| 2 | 2.006315 | -0.297524 | 0 |
| 3 | 0.077145 | 0.684294 | 3 |
| 4 | -0.385682 | -1.611826 | 2 |
# Show cluster counts
print(df_kmeans["cluster"].value_counts().sort_index())
0 125 1 125 2 125 3 125 Name: cluster, dtype: int64
📊 Visualize Clusters¶
def plot_clusters_2d(df, label_col="cluster", centroids=None, title="Cluster Visualization (2D)", hue_palette="tab10"):
"""
Plots a 2D scatterplot of clusters with optional centroids.
Parameters:
df (pd.DataFrame): DataFrame with exactly 2 feature columns and a cluster label column.
label_col (str): Name of the column containing cluster labels.
centroids (ndarray or list): Optional array of centroid coordinates (shape: [K, 2]).
title (str): Plot title.
hue_palette (str): Color palette for clusters.
"""
plt.figure(figsize=(6, 5))
sns.scatterplot(data=df, x=df.columns[0], y=df.columns[1], hue=label_col, palette=hue_palette, s=50, edgecolor="k")
if centroids is not None:
plt.scatter(centroids[:, 0], centroids[:, 1], c='black', s=150, marker='X', label='Centroids')
plt.title(title)
plt.legend()
plt.show()
plot_clusters_2d(df_kmeans, label_col="cluster", centroids=kmeans_final.cluster_centers_, title=f"KMeans Clustering (K = {k_final})")
from sklearn.decomposition import PCA
import matplotlib.pyplot as plt
import seaborn as sns
def plot_clusters_pca(df_with_labels, label_col="cluster", title="Cluster Visualization (PCA-backed)"):
"""
Plots clusters in 2D using PCA if necessary.
Parameters:
df_with_labels (pd.DataFrame): DataFrame with features + cluster label column.
label_col (str): Name of the cluster label column.
title (str): Plot title.
"""
features = df_with_labels.drop(columns=[label_col])
# Reduce to 2D if needed
if features.shape[1] > 2:
pca = PCA(n_components=2, random_state=42)
reduced = pca.fit_transform(features)
plot_df = pd.DataFrame(reduced, columns=["PC1", "PC2"])
else:
plot_df = features.copy()
plot_df.columns = ["PC1", "PC2"]
plot_df[label_col] = df_with_labels[label_col].values
plt.figure(figsize=(6, 5))
sns.scatterplot(data=plot_df, x="PC1", y="PC2", hue=label_col, palette="tab10", s=50, edgecolor="k")
plt.title(title)
plt.show()
plot_clusters_pca(df_kmeans, label_col="cluster", title=f"KMeans Clustering (K = {k_final})")
📌 Cluster Summary¶
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
def summarize_clusters(df, label_col="cluster", summary_cols=None, cmap="Blues"):
"""
Displays summary statistics for each cluster.
Parameters:
df (pd.DataFrame): DataFrame with features + cluster label.
label_col (str): Column name containing cluster labels.
summary_cols (list): List of columns to summarize. If None, all numeric features are used.
cmap (str): Color map for background gradient.
"""
if summary_cols is None:
summary_cols = [col for col in df.select_dtypes(include="number").columns if col != label_col]
summary = df.groupby(label_col).agg(
count=(label_col, "size"),
**{f"avg_{col}": (col, "mean") for col in summary_cols}
).reset_index()
return summary.style.background_gradient(cmap=cmap, axis=0)
summarize_clusters(df_kmeans)
| cluster | count | avg_feature_1 | avg_feature_2 | |
|---|---|---|---|---|
| 0 | 0 | 125 | 1.530125 | -0.154559 |
| 1 | 1 | 125 | -0.990375 | 0.719419 |
| 2 | 2 | 125 | -0.684463 | -1.539546 |
| 3 | 3 | 125 | 0.144713 | 0.974686 |
🧱 Hierarchical Clustering¶
📖 Click to Expand
🧱 What is Hierarchical Clustering?¶
Hierarchical clustering builds a nested tree of clusters by either:
- Agglomerative (bottom-up): Merge closest clusters until one remains
- Divisive (top-down): Start with one big cluster and recursively split
It does not require you to predefine the number of clusters — instead, you choose where to “cut” the tree.
🔹 Distance Between Clusters (Linkage Types)¶
- Single linkage: Minimum distance between points in two clusters
- Complete linkage: Maximum distance
- Average linkage: Mean pairwise distance
- Ward linkage: Minimizes increase in total variance (best for compact clusters)
✅ When to Use¶
- You want a visual tree of how data groups form
- You’re unsure how many clusters to choose — let the tree reveal it
- You want interpretable cluster evolution (e.g., customer groups merging)
⚠️ Limitations¶
- Memory and time intensive for large datasets
- Sensitive to scale and noise
- Tree depth can be misleading without proper distance normalization
⚙️ Hierarchical Config¶
from sklearn.preprocessing import StandardScaler
# === CONFIG ===
hierarchical_metric = "euclidean" # Distance metric (euclidean, manhattan, etc.)
hierarchical_linkage = "ward" # Linkage: ward, single, complete, average
n_clusters_hierarchical = 4 # Number of clusters to cut the tree at
use_scaled_data_hierarchical = True # Whether to scale features
# Prepare data
df_input_hier = df.copy()
if use_scaled_data_hierarchical:
scaler_hier = StandardScaler()
df_input_hier = pd.DataFrame(scaler_hier.fit_transform(df), columns=df.columns)
df_input_hier.head()
| feature_1 | feature_2 | |
|---|---|---|
| 0 | -0.766161 | 0.562117 |
| 1 | -1.266310 | -1.545187 |
| 2 | 2.006315 | -0.297524 |
| 3 | 0.077145 | 0.684294 |
| 4 | -0.385682 | -1.611826 |
🌳 Plot Dendrogram¶
📖 Click to Expand
🌳 What is a Dendrogram?¶
A dendrogram is a tree diagram that shows how points are merged into clusters during agglomerative clustering.
- X-axis: data points or their indexes
- Y-axis: distance between merged clusters
- You can “cut” the tree at any height to form flat clusters
🔍 Why It’s Useful¶
- Visualizes the entire clustering process, not just final clusters
- Helps choose the optimal number of clusters based on vertical gaps
- Useful when you don’t know how many clusters to use up front
from scipy.cluster.hierarchy import dendrogram, linkage
import matplotlib.pyplot as plt
# Compute linkage matrix
linkage_matrix = linkage(df_input_hier, method=hierarchical_linkage, metric=hierarchical_metric)
# Plot dendrogram
plt.figure(figsize=(10, 4))
dendrogram(linkage_matrix, truncate_mode='level', p=10)
plt.title(f"Hierarchical Dendrogram ({hierarchical_linkage.title()} Linkage)")
plt.xlabel("Data Points")
plt.ylabel(f"{hierarchical_metric.title()} Distance")
plt.show()
✂️ Choose Clusters from Tree¶
📖 Click to Expand
✂️ Cutting the Dendrogram into Flat Clusters¶
Once the hierarchical tree (dendrogram) is built, we flatten it into K clusters using a cutting rule.
🔧 How It Works¶
- The dendrogram represents how clusters were merged based on distance
- We choose a number
K, and cut the tree horizontally to extract K flat clusters - Internally, this is done using:
criterion='maxclust': Find the highest level whereKclusters existt=K: The number of clusters to form
🧠 Key Benefit¶
- Lets you defer the choice of K until after seeing the full tree
- Supports exploratory workflows (e.g., try K = 3, 4, 5 and compare)
⚠️ Notes¶
- If clusters are very unbalanced or chaining happens, some clusters may be very small
- A good “cut height” usually corresponds to large vertical gaps in the dendrogram
# Recompute linkage_matrix if not already defined
from scipy.cluster.hierarchy import linkage, fcluster
linkage_matrix = linkage(df_input_hier, method=hierarchical_linkage, metric=hierarchical_metric)
# Cut the tree
labels_hier = fcluster(linkage_matrix, t=n_clusters_hierarchical, criterion='maxclust')
# Add to dataframe
df_hier = df_input_hier.copy()
df_hier["cluster"] = labels_hier
# Cluster counts
df_hier["cluster"].value_counts().sort_index()
1 125 2 125 3 126 4 124 Name: cluster, dtype: int64
📊 Visualize Clusters (PCA-backed)¶
plot_clusters_pca(df_hier, label_col="cluster", title="Hierarchical Clustering (PCA-backed)")
plot_clusters_2d(df_hier, label_col="cluster", title="Hierarchical Clustering (2D)")
📌 Cluster Summary¶
summarize_clusters(df_hier)
| cluster | count | avg_feature_1 | avg_feature_2 | |
|---|---|---|---|---|
| 0 | 1 | 125 | -0.684463 | -1.539546 |
| 1 | 2 | 125 | 1.530125 | -0.154559 |
| 2 | 3 | 126 | -0.985897 | 0.722557 |
| 3 | 4 | 124 | 0.149316 | 0.973557 |
🌐 DBSCAN¶
🌐 DBSCAN¶
📖 Click to Expand
🌐 What is DBSCAN?¶
DBSCAN (Density-Based Spatial Clustering of Applications with Noise) groups points into clusters based on density.
- Points in high-density areas → core points
- Border points are near core points
- Noise points are too isolated to join any cluster
🔧 Key Parameters¶
eps: Radius for neighborhood searchmin_samples: Minimum neighbors to form a dense region
✅ When to Use¶
- You expect irregularly shaped clusters
- You want to automatically detect outliers
- No need to specify
Kupfront
⚠️ Limitations¶
- Requires careful tuning of
eps - Sensitive to feature scaling
- Struggles when density varies too much across regions
⚙️ DBSCAN Config¶
from sklearn.preprocessing import StandardScaler
# === CONFIG ===
dbscan_eps = 0.5 # Radius for neighbors
dbscan_min_samples = 5 # Minimum points to form a dense region
use_scaled_data_dbscan = True
# Prepare input
df_input_dbscan = df.copy()
if use_scaled_data_dbscan:
scaler_db = StandardScaler()
df_input_dbscan = pd.DataFrame(scaler_db.fit_transform(df), columns=df.columns)
🚀 Run DBSCAN¶
from sklearn.cluster import DBSCAN
# Fit DBSCAN
dbscan = DBSCAN(eps=dbscan_eps, min_samples=dbscan_min_samples)
labels_dbscan = dbscan.fit_predict(df_input_dbscan)
# Assign labels
df_dbscan = df_input_dbscan.copy()
df_dbscan["cluster"] = labels_dbscan
df_dbscan
| feature_1 | feature_2 | cluster | |
|---|---|---|---|
| 0 | -0.766161 | 0.562117 | 0 |
| 1 | -1.266310 | -1.545187 | 1 |
| 2 | 2.006315 | -0.297524 | 2 |
| 3 | 0.077145 | 0.684294 | 0 |
| 4 | -0.385682 | -1.611826 | 1 |
| ... | ... | ... | ... |
| 495 | -0.709665 | 0.868735 | 0 |
| 496 | 0.148308 | 1.024226 | 0 |
| 497 | -0.732909 | -1.220579 | 1 |
| 498 | -0.775140 | -1.337366 | 1 |
| 499 | 1.628939 | 0.143440 | 2 |
500 rows × 3 columns
📊 Visual Output¶
plot_clusters_2d(df_dbscan, label_col="cluster", title="DBSCAN Clustering (2D)")
plot_clusters_pca(df_dbscan, label_col="cluster", title="DBSCAN Clustering (PCA-backed)")
📌 Cluster Summary¶
summarize_clusters(df_dbscan)
| cluster | count | avg_feature_1 | avg_feature_2 | |
|---|---|---|---|---|
| 0 | 0 | 250 | -0.422831 | 0.847053 |
| 1 | 1 | 125 | -0.684463 | -1.539546 |
| 2 | 2 | 125 | 1.530125 | -0.154559 |
🎲 Gaussian Mixture Models (GMM)¶
🎲 Gaussian Mixture Models (GMM)¶
📖 Click to Expand
🎲 What is GMM?¶
GMM is a probabilistic clustering method that assumes data is generated from a mixture of several Gaussian distributions.
- Each cluster is a Gaussian component defined by its own mean and covariance
- Unlike KMeans, it produces soft assignments (probabilities)
✅ When to Use¶
- Clusters may overlap and have elliptical shapes
- You need probabilistic membership, not just hard labels
- Ideal for continuous numeric features
⚙️ Key Parameters¶
n_components: Number of clusterscovariance_type: Full, tied, diagonal, sphericalrandom_state: For reproducibility
⚠️ Limitations¶
- Assumes Gaussian-shaped clusters
- Sensitive to initial guesses and scaling
- Can overfit if too many components
⚙️ GMM Config¶
# === CONFIG ===
gmm_components = 4
gmm_covariance_type = "full"
use_scaled_data_gmm = True
# Prepare input
df_input_gmm = df.copy()
if use_scaled_data_gmm:
scaler_gmm = StandardScaler()
df_input_gmm = pd.DataFrame(scaler_gmm.fit_transform(df), columns=df.columns)
🚀 Run GMM¶
from sklearn.mixture import GaussianMixture
# Fit GMM
gmm = GaussianMixture(n_components=gmm_components, covariance_type=gmm_covariance_type, random_state=42)
labels_gmm = gmm.fit_predict(df_input_gmm)
# Assign labels
df_gmm = df_input_gmm.copy()
df_gmm["cluster"] = labels_gmm
📊 Visualize Clusters (PCA-backed)¶
plot_clusters_pca(df_gmm, label_col="cluster", title="GMM Clustering (PCA-backed)")
plot_clusters_2d(df_gmm, label_col="cluster", title="GMM Clustering (2D)")
📌 Cluster Summary¶
summarize_clusters(df_gmm)
| cluster | count | avg_feature_1 | avg_feature_2 | |
|---|---|---|---|---|
| 0 | 0 | 125 | 1.530125 | -0.154559 |
| 1 | 1 | 125 | -0.990375 | 0.719419 |
| 2 | 2 | 125 | -0.684463 | -1.539546 |
| 3 | 3 | 125 | 0.144713 | 0.974686 |
📍 Mean Shift¶
📍 Mean Shift¶
📖 Click to Expand
📍 What is Mean Shift?¶
Mean Shift is a centroid-based clustering algorithm that locates high-density regions (modes) by shifting points toward the mean of their local neighborhood.
- No need to specify number of clusters (
K) - Each data point moves to the densest area until convergence
✅ When to Use¶
- You want automatic cluster count detection
- Data has dense blobs or modes
- Good for small-to-medium datasets
⚙️ Key Parameters¶
bandwidth: Radius of the neighborhood (if not provided, it's estimated)bin_seeding: Speed optimization using binning
⚠️ Limitations¶
- Computationally expensive
- Highly sensitive to bandwidth
- Doesn’t scale well for high dimensions
⚙️ Mean Shift Config¶
use_scaled_data_meanshift = True
# Prepare input
df_input_meanshift = df.copy()
if use_scaled_data_meanshift:
scaler_ms = StandardScaler()
df_input_meanshift = pd.DataFrame(scaler_ms.fit_transform(df), columns=df.columns)
🚀 Run Mean Shift¶
from sklearn.cluster import MeanShift, estimate_bandwidth
# Estimate bandwidth
bandwidth = estimate_bandwidth(df_input_meanshift, quantile=0.2, n_samples=200)
# Fit MeanShift
meanshift = MeanShift(bandwidth=bandwidth, bin_seeding=True)
labels_meanshift = meanshift.fit_predict(df_input_meanshift)
# Assign labels
df_meanshift = df_input_meanshift.copy()
df_meanshift["cluster"] = labels_meanshift
📊 Visualize Clusters (PCA-backed)¶
plot_clusters_pca(df_meanshift, label_col="cluster", title="Mean Shift Clustering (PCA-backed)")
# or if 2D:
plot_clusters_2d(df_meanshift, label_col="cluster", title="Mean Shift Clustering (2D)")
📌 Cluster Summary¶
summarize_clusters(df_meanshift)
| cluster | count | avg_feature_1 | avg_feature_2 | |
|---|---|---|---|---|
| 0 | 0 | 125 | -0.990375 | 0.719419 |
| 1 | 1 | 125 | 1.530125 | -0.154559 |
| 2 | 2 | 125 | 0.144713 | 0.974686 |
| 3 | 3 | 125 | -0.684463 | -1.539546 |
🎼 Spectral Clustering¶
📖 Click to Expand
🎼 What is Spectral Clustering?¶
Spectral clustering transforms the data into a graph of similarities, then finds clusters by analyzing the eigenvectors of the graph Laplacian.
- Works well for non-convex and irregular cluster shapes
- Doesn’t assume globular structure like KMeans
✅ When to Use¶
- When data has nonlinear structure (e.g., concentric circles)
- For small-to-medium datasets
- When traditional clustering fails on irregular shapes
⚙️ Key Parameters¶
n_clusters: Number of output clustersaffinity: Similarity measure (e.g.,'rbf','nearest_neighbors')assign_labels: Method to assign final clusters ('kmeans','discretize')
⚠️ Limitations¶
- Slow for large datasets (due to eigen decomposition)
- Needs tuning of affinity or neighbors
- Doesn't scale well beyond a few thousand points
⚙️ Spectral Config¶
spectral_clusters = 4
spectral_affinity = "rbf" # or 'nearest_neighbors'
use_scaled_data_spectral = True
# Prepare input
df_input_spectral = df.copy()
if use_scaled_data_spectral:
scaler_spectral = StandardScaler()
df_input_spectral = pd.DataFrame(scaler_spectral.fit_transform(df), columns=df.columns)
🚀 Run Spectral Clustering¶
from sklearn.cluster import SpectralClustering
# Fit model
spectral = SpectralClustering(n_clusters=spectral_clusters,
affinity=spectral_affinity,
assign_labels="kmeans",
random_state=42)
labels_spectral = spectral.fit_predict(df_input_spectral)
# Assign labels
df_spectral = df_input_spectral.copy()
df_spectral["cluster"] = labels_spectral
📊 Visualize Clusters (PCA-backed)¶
plot_clusters_pca(df_spectral, label_col="cluster", title="Spectral Clustering (PCA-backed)")
# or if 2D:
plot_clusters_2d(df_spectral, label_col="cluster", title="Spectral Clustering (2D)")
📌 Cluster Summary¶
summarize_clusters(df_spectral)
| cluster | count | avg_feature_1 | avg_feature_2 | |
|---|---|---|---|---|
| 0 | 0 | 125 | -0.684463 | -1.539546 |
| 1 | 1 | 125 | 0.144713 | 0.974686 |
| 2 | 2 | 125 | 1.530125 | -0.154559 |
| 3 | 3 | 125 | -0.990375 | 0.719419 |
📡 OPTICS¶
📡 OPTICS¶
📖 Click to Expand
📡 What is OPTICS?¶
OPTICS (Ordering Points To Identify the Clustering Structure) is a density-based clustering algorithm similar to DBSCAN but more robust.
- Unlike DBSCAN, OPTICS doesn’t force you to set a fixed
epsradius - It builds a reachability plot to identify clusters of varying density
✅ When to Use¶
- When DBSCAN fails due to mixed-density clusters
- You want to extract nested or chained cluster structures
⚙️ Key Parameters¶
min_samples: Minimum points in a neighborhood to form a core pointxi: Sensitivity for extracting flat clusters from reachability structure
⚠️ Limitations¶
- Slower than DBSCAN for large datasets
- Still needs thoughtful tuning of
min_samplesandxi
⚙️ OPTICS Config¶
optics_min_samples = 5
optics_xi = 0.05
use_scaled_data_optics = True
df_input_optics = df.copy()
if use_scaled_data_optics:
scaler_optics = StandardScaler()
df_input_optics = pd.DataFrame(scaler_optics.fit_transform(df), columns=df.columns)
🚀 Run OPTICS¶
📖 Click to Expand
🔁 How OPTICS Works¶
- For each point, compute the core distance: the minimum radius to have
min_samplesneighbors - Build a reachability graph:
- A reachability distance is defined for each pair based on core distances
- Use a priority queue to order points by how reachable they are from a known cluster
- Extract clusters from the reachability plot using
xi— a steep drop indicates a cluster boundary
from sklearn.cluster import OPTICS
# Fit OPTICS
optics = OPTICS(min_samples=optics_min_samples, xi=optics_xi)
labels_optics = optics.fit_predict(df_input_optics)
# Assign labels
df_optics = df_input_optics.copy()
df_optics["cluster"] = labels_optics
📊 Visualize Clusters (PCA-backed)¶
plot_clusters_pca(df_optics, label_col="cluster", title="OPTICS Clustering (PCA-backed)")
# or if 2D:
plot_clusters_2d(df_optics, label_col="cluster", title="OPTICS Clustering (2D)")
📌 Cluster Summary¶
summarize_clusters(df_optics)
| cluster | count | avg_feature_1 | avg_feature_2 | |
|---|---|---|---|---|
| 0 | -1 | 311 | -0.004814 | -0.082243 |
| 1 | 0 | 10 | -0.959704 | 0.602515 |
| 2 | 1 | 5 | -1.162292 | 0.619093 |
| 3 | 2 | 15 | -1.087775 | 0.741034 |
| 4 | 3 | 6 | -0.863286 | 0.770533 |
| 5 | 4 | 7 | -1.030488 | 0.954377 |
| 6 | 5 | 5 | -1.001313 | 0.422589 |
| 7 | 6 | 6 | -0.074199 | 1.037078 |
| 8 | 7 | 10 | 0.002344 | 0.897399 |
| 9 | 8 | 17 | 0.211347 | 1.074383 |
| 10 | 9 | 8 | 0.196804 | 0.872040 |
| 11 | 10 | 6 | 0.179127 | 0.736518 |
| 12 | 11 | 5 | -0.173858 | 0.842246 |
| 13 | 12 | 6 | 0.589280 | 1.038262 |
| 14 | 13 | 7 | 1.314370 | -0.071212 |
| 15 | 14 | 9 | 1.325220 | -0.211189 |
| 16 | 15 | 6 | 1.515064 | -0.307475 |
| 17 | 16 | 6 | 1.589649 | -0.283244 |
| 18 | 17 | 7 | 1.505977 | -0.038246 |
| 19 | 18 | 5 | 1.482161 | 0.071092 |
| 20 | 19 | 6 | 1.664769 | -0.012977 |
| 21 | 20 | 6 | -0.629999 | -1.364548 |
| 22 | 21 | 13 | -0.598810 | -1.582801 |
| 23 | 22 | 6 | -0.714511 | -1.598947 |
| 24 | 23 | 7 | -0.788284 | -1.424303 |
| 25 | 24 | 5 | -0.838434 | -1.822272 |
🌲 BIRCH¶
🌲 BIRCH¶
📖 Click to Expand
🌲 What is BIRCH?¶
BIRCH (Balanced Iterative Reducing and Clustering using Hierarchies) is a scalable clustering method designed for large datasets.
- It builds a Clustering Feature Tree (CF Tree) incrementally
- Then performs final clustering (e.g., via KMeans) on condensed representations
✅ When to Use¶
- Very large datasets (millions of points)
- You want online, memory-efficient clustering
- Data has compact, globular clusters
⚙️ Key Parameters¶
threshold: Max radius for subclusters in the CF treebranching_factor: Max children per node in the treen_clusters: Optional number of final clusters
⚠️ Limitations¶
- Assumes clusters are spherical and evenly sized
- Not great for non-convex shapes or overlapping densities
⚙️ BIRCH Config¶
birch_threshold = 0.5
birch_n_clusters = 4
use_scaled_data_birch = True
df_input_birch = df.copy()
if use_scaled_data_birch:
scaler_birch = StandardScaler()
df_input_birch = pd.DataFrame(scaler_birch.fit_transform(df), columns=df.columns)
🚀 Run BIRCH¶
📖 Click to Expand
🔁 How BIRCH Works¶
- Build a CF Tree:
- Summarizes incoming data into compact representations
- Each node stores statistics (count, linear sum, square sum)
- Condense:
- Leaf nodes represent microclusters (summary objects)
- Cluster:
- Apply global clustering (e.g., KMeans) to the leaf entries
BIRCH is ideal for incremental learning and can cluster data without full memory loading.
from sklearn.cluster import Birch
birch = Birch(threshold=birch_threshold, n_clusters=birch_n_clusters)
labels_birch = birch.fit_predict(df_input_birch)
df_birch = df_input_birch.copy()
df_birch["cluster"] = labels_birch
📊 Visualize Clusters (PCA-backed)¶
plot_clusters_pca(df_birch, label_col="cluster", title="BIRCH Clustering (PCA-backed)")
# or if 2D:
plot_clusters_2d(df_birch, label_col="cluster", title="BIRCH Clustering (2D)")
📌 Cluster Summary¶
summarize_clusters(df_birch)
| cluster | count | avg_feature_1 | avg_feature_2 | |
|---|---|---|---|---|
| 0 | 0 | 127 | 0.135329 | 0.969345 |
| 1 | 1 | 123 | -0.999143 | 0.720783 |
| 2 | 2 | 125 | 1.530125 | -0.154559 |
| 3 | 3 | 125 | -0.684463 | -1.539546 |
🔥 HDBSCAN¶
📖 Click to Expand
🔥 What is HDBSCAN?¶
HDBSCAN (Hierarchical Density-Based Spatial Clustering of Applications with Noise) is a density-based clustering method that extends DBSCAN with:
- Variable density support
- Hierarchical clustering of core points
- Automatic cluster selection using stability scores
It is more flexible than DBSCAN, especially in complex datasets.
✅ When to Use¶
- Data has clusters with varying densities
- You want automatic cluster count detection
- DBSCAN fails due to fixed-radius limitations
⚙️ Key Parameters¶
min_cluster_size: Minimum number of samples per clustermin_samples: Density threshold (can be left None)cluster_selection_epsilon: Forces tighter clustering
⚠️ Limitations¶
- Slower than DBSCAN
- Output cluster labels may skip numbers (e.g., 0, 1, 5)
⚙️ HDBSCAN Config¶
use_scaled_data_hdbscan = True
min_cluster_size_hdbscan = 10
min_samples_hdbscan = None # Optional
df_input_hdbscan = df.copy()
if use_scaled_data_hdbscan:
scaler_hdbscan = StandardScaler()
df_input_hdbscan = pd.DataFrame(scaler_hdbscan.fit_transform(df), columns=df.columns)
🚀 Run HDBSCAN¶
📖 Click to Expand
🔁 How HDBSCAN Works¶
- Estimate mutual reachability distances
- Build a minimum spanning tree (MST) from core distances
- Perform hierarchical clustering from the MST
- Extract clusters using stability scores — points that persist across resolutions are considered stable
# !pip install hdbscan
import hdbscan
hdb = hdbscan.HDBSCAN(min_cluster_size=min_cluster_size_hdbscan,
min_samples=min_samples_hdbscan)
labels_hdbscan = hdb.fit_predict(df_input_hdbscan)
df_hdbscan = df_input_hdbscan.copy()
df_hdbscan["cluster"] = labels_hdbscan
📊 Visualize Clusters (PCA-backed)¶
plot_clusters_pca(df_hdbscan, label_col="cluster", title="HDBSCAN Clustering (PCA-backed)")
# or if 2D:
plot_clusters_2d(df_hdbscan, label_col="cluster", title="HDBSCAN Clustering (2D)")
📌 Cluster Summary¶
summarize_clusters(df_hdbscan)
| cluster | count | avg_feature_1 | avg_feature_2 | |
|---|---|---|---|---|
| 0 | -1 | 1 | 0.282082 | 1.685677 |
| 1 | 0 | 125 | -0.684463 | -1.539546 |
| 2 | 1 | 125 | 1.530125 | -0.154559 |
| 3 | 2 | 125 | -0.990375 | 0.719419 |
| 4 | 3 | 124 | 0.143605 | 0.968953 |
🧬 Affinity Propagation¶
📖 Click to Expand
🧬 What is Affinity Propagation?¶
Affinity Propagation is a clustering algorithm that identifies exemplars (representative points) based on message-passing between data points.
- It automatically determines the number of clusters
- It builds clusters around the most “influential” samples
✅ When to Use¶
- You want automatic cluster count
- You prefer representative samples (exemplars) over centroids
- Dataset is not too large (O(N²) memory)
⚙️ Key Parameters¶
preference: Controls number of clusters (lower = more clusters)damping: Smoothing factor between iterationsaffinity: Similarity metric (default is negative squared Euclidean)
⚠️ Limitations¶
- Slow and memory-heavy for large datasets
- Sensitive to
preferencetuning - Doesn’t scale well beyond a few thousand samples
⚙️ Affinity Config¶
affinity_preference = None # Optional: controls number of clusters
use_scaled_data_affinity = True
df_input_affinity = df.copy()
if use_scaled_data_affinity:
scaler_aff = StandardScaler()
df_input_affinity = pd.DataFrame(scaler_aff.fit_transform(df), columns=df.columns)
🚀 Run Affinity Propagation¶
📖 Click to Expand
🔁 How Affinity Propagation Works¶
- All points send “responsibility” messages: how well they'd represent others
- All points receive “availability” messages: how good others are at representing them
- Points with high combined score become exemplars
- Others are assigned to the closest exemplar
The method iteratively updates these messages until convergence.
from sklearn.cluster import AffinityPropagation
aff = AffinityPropagation(preference=affinity_preference, random_state=42)
labels_affinity = aff.fit_predict(df_input_affinity)
df_affinity = df_input_affinity.copy()
df_affinity["cluster"] = labels_affinity
/Users/ashrithreddy/anaconda3/lib/python3.11/site-packages/sklearn/cluster/_affinity_propagation.py:143: ConvergenceWarning: Affinity propagation did not converge, this model may return degenerate cluster centers and labels. warnings.warn(
📊 Visualize Clusters (PCA-backed)¶
plot_clusters_pca(df_affinity, label_col="cluster", title="Affinity Propagation (PCA-backed)")
# or if 2D:
plot_clusters_2d(df_affinity, label_col="cluster", title="Affinity Propagation (2D)")
📌 Cluster Summary¶
summarize_clusters(df_affinity)
| cluster | count | avg_feature_1 | avg_feature_2 | |
|---|---|---|---|---|
| 0 | 0 | 54 | 1.747547 | -0.147736 |
| 1 | 1 | 32 | -0.043576 | 1.077206 |
| 2 | 2 | 71 | 1.364762 | -0.159749 |
| 3 | 3 | 21 | -0.055143 | 0.826704 |
| 4 | 4 | 32 | 0.208257 | 0.806212 |
| 5 | 5 | 40 | 0.349433 | 1.105142 |
| 6 | 6 | 67 | -1.109341 | 0.814806 |
| 7 | 7 | 77 | -0.609241 | -1.651268 |
| 8 | 8 | 58 | -0.852950 | 0.609232 |
| 9 | 9 | 48 | -0.805131 | -1.360326 |
📌 Summary Table¶
📋 Comparison Across Methods¶
# Re-run without stray markdown syntax error
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.decomposition import PCA
def plot_all_cluster_outputs(
clustered_dfs,
label_cols=None,
method="auto",
title_prefix=""
):
"""
Plots clustering results from multiple algorithms on the same dataset using either:
- raw 2D features,
- PCA-reduced 2D,
- or auto mode (PCA if >2 features).
Parameters:
clustered_dfs (dict): Dictionary of {method_name: df_with_cluster_labels}
label_cols (dict): Optional dictionary {method_name: label_col_name}. If None, defaults to 'cluster'
method (str or list): "pca", "auto", or list of [col1, col2] for raw plotting
title_prefix (str): Optional prefix for each subplot title
"""
n_methods = len(clustered_dfs)
fig, axes = plt.subplots(1, n_methods, figsize=(4 * n_methods, 4), squeeze=False)
for idx, (name, df_clustered) in enumerate(clustered_dfs.items()):
label_col = label_cols.get(name, "cluster") if label_cols else "cluster"
features = df_clustered.drop(columns=[label_col])
# Determine what to plot
if isinstance(method, list) and len(method) == 2:
plot_df = df_clustered[method].copy()
plot_df.columns = ["X", "Y"]
elif method == "pca" or (method == "auto" and features.shape[1] > 2):
pca = PCA(n_components=2, random_state=42)
reduced = pca.fit_transform(features)
plot_df = pd.DataFrame(reduced, columns=["X", "Y"])
else: # raw features
plot_df = features.iloc[:, :2].copy()
plot_df.columns = ["X", "Y"]
plot_df[label_col] = df_clustered[label_col].values
ax = axes[0, idx]
sns.scatterplot(data=plot_df, x="X", y="Y", hue=label_col, palette="tab10", s=10, edgecolor=None, ax=ax)
ax.set_title(f"{title_prefix}{name}", fontsize=10)
ax.set_xlabel("")
ax.set_ylabel("")
ax.set_xticks([])
ax.set_yticks([])
ax.legend().remove()
plt.tight_layout()
plt.show()
plot_all_cluster_outputs(
clustered_dfs={
"KMeans": df_kmeans,
"Hierarchical": df_hier,
"DBSCAN": df_dbscan,
"GMM": df_gmm,
"Mean Shift": df_meanshift,
"Spectral": df_spectral,
"OPTICS": df_optics,
"BIRCH": df_birch,
"HDBSCAN": df_hdbscan,
"Affinity Propagation": df_affinity
},
label_cols={
"KMeans": "cluster",
"Hierarchical": "cluster",
"DBSCAN": "cluster",
"GMM": "cluster",
"Mean Shift": "cluster",
"Spectral": "cluster",
"OPTICS": "cluster",
"BIRCH": "cluster",
"HDBSCAN": "cluster",
"Affinity Propagation": "cluster"
},
method="pca", # or "auto", or ["feature_1", "feature_2"]
title_prefix=""
)
🧭 Practical Recommendations¶
🧭 Click to Expand
Q1: Do you know how many clusters you need?
├─ Yes →
├─ Are clusters roughly spherical and well-separated?
├─ Yes → ✅ Use KMeans
└─ No → ✅ Use GMM (soft boundaries)
└─ No →
├─ Do you expect clusters of varying density or arbitrary shape?
├─ Yes →
├─ Is your dataset small or moderate in size?
├─ Yes → ✅ Use HDBSCAN
└─ No → ✅ Use OPTICS
├─ No → ✅ Use Mean Shift or Affinity Propagation
Q2: Is your dataset graph-structured or non-linearly separable?
└─ Yes → ✅ Use Spectral Clustering
Q3: Is your dataset large (millions of points)?
└─ Yes → ✅ Use BIRCH



